|
||
|
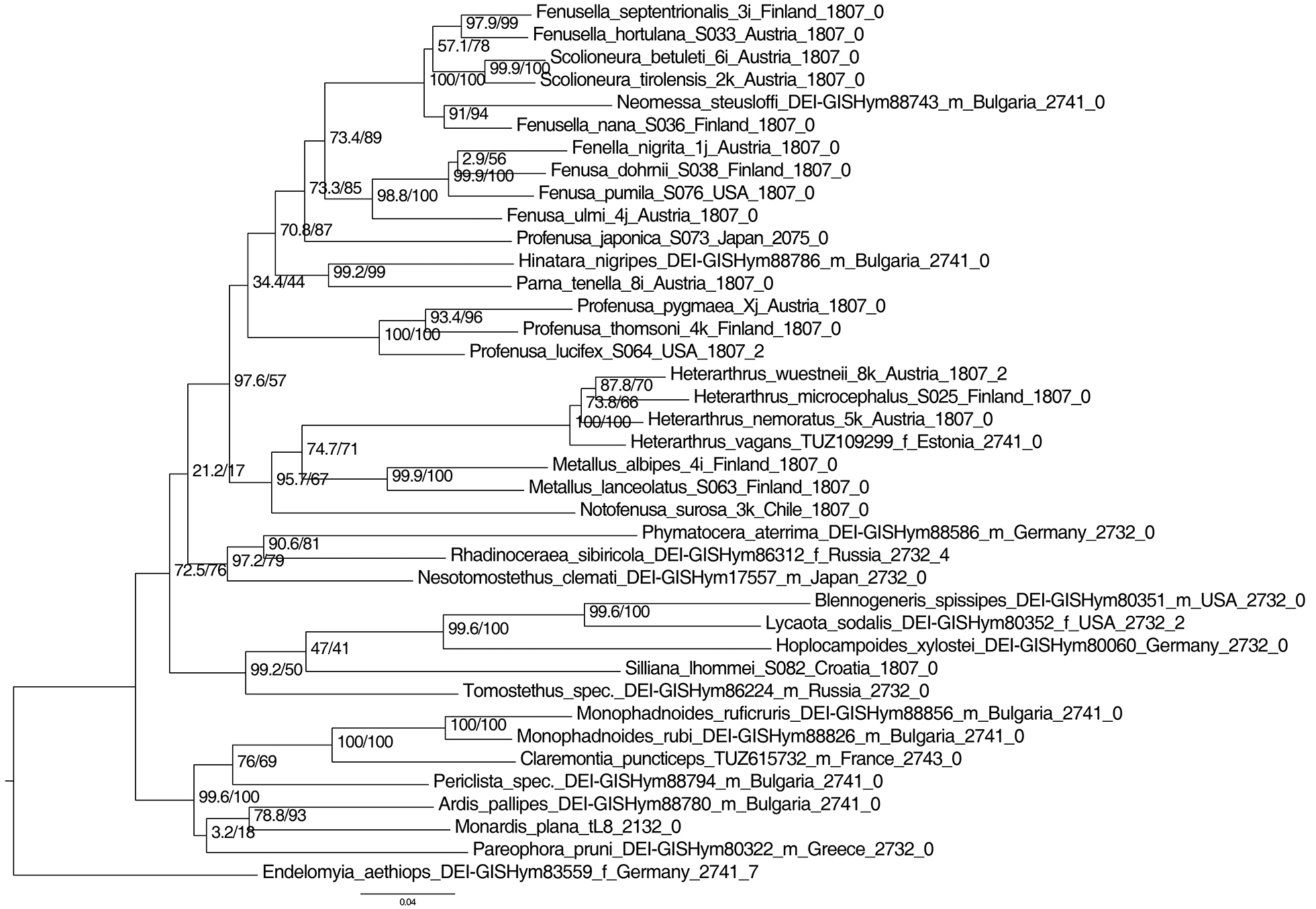
Maximum likelihood tree of Blennocampinae and Heterarthrinae based on two genes (COI and NaK). Best-fit model chosen according to Bayesian information criterion was GTR+I+G4. Numbers beside nodes show SH-aLRT support (%) / ultrafast bootstrap support (%) values. Support values for weakly supported branches (<90) are not shown. Letters “f” and “m” stand for “female” and “male”. Numbers at the end of the tip labels refer to the length of the sequence and the number of ambiguous positions (e.g. polymorphisms). The tree was rooted according to the results of Leppänen et al. (2012). The scale bar shows the number of estimated substitutions per nucleotide position. |