|
||
|
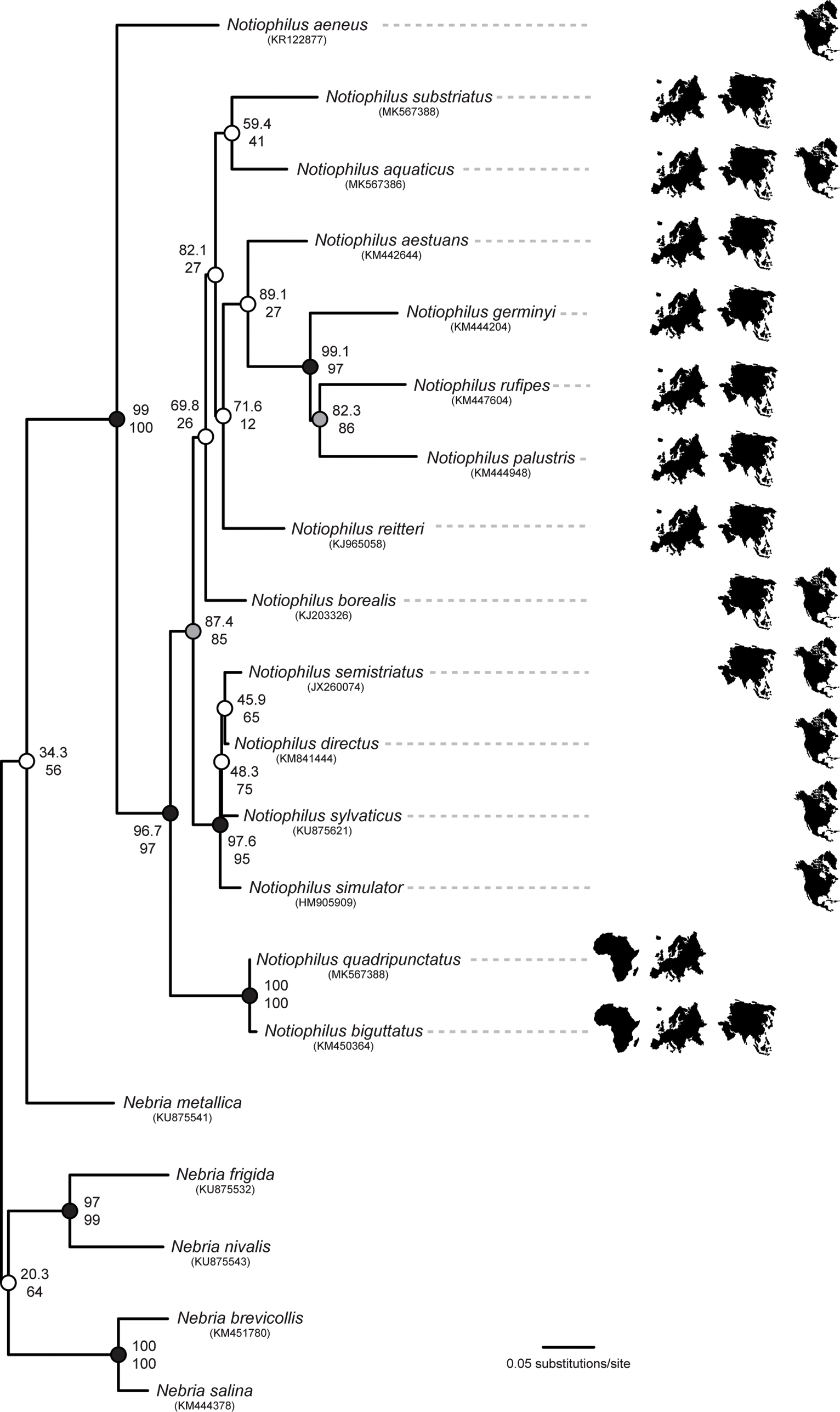
Maximum likelihood phylogeny inferred in IQ-TREE, based on the CO1 barcode fragment for the genus Notiophilus. The model of nucleotide substitution used was selected with Modelfinder as part of the IQ-TREE work package. The tree was rooted with five Nebria species as outgroup. Nodal support was calculated with SH-aLRT (above) and UFBoot (below) values. Black dots indicate very robust nodes with very high values (SH-aLRT ≥ 90%, UFBoot ≥ 95%), grey dots indicate moderately robust nodes (SH-aLRT ≥ 80%, UFBoot ≥ 80%) and white dots indicate weak nodes (SH-aLRT < 80%, UFBoot < 80%) (see Material and Methods for details). Continent silhouettes indicate the biogeographic distribution of the analysed taxa (from left to right: Africa, Europe, Asia and North America). |